hankyoreh
Links to other country sites 다른 나라 사이트 링크
S. Korean research team compiles high-resolution genetic map of novel coronavirus
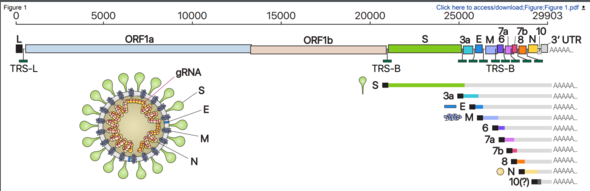
In a joint study with the Korea National Institute of Health, under the Korea Centers for Disease Control and Prevention (KCDC), the Center for RNA Research at the Institute for Basic Science (IBS) has managed to compile a high-resolution genetic map based on an analysis of the transcriptome of the RNA created in host cells by SARS-CoV-2, the novel coronavirus that causes the disease COVID-19. The researchers discovered dozens of previously unknown types of RNA and at least 41 RNA modification sites that present possible targets for medication being developed to treat COVID-19.
A research team led by V. Narry Kim, director of the center, and Chang Hye-shik, professor of biological sciences at Seoul National University, analyzed all the base sequences in the transcriptome (set of all RNA molecules) that SARS-CoV-2 creates in its host cells. Through this analysis, they were able to determine with accuracy the location of the genes in genomic RNA.
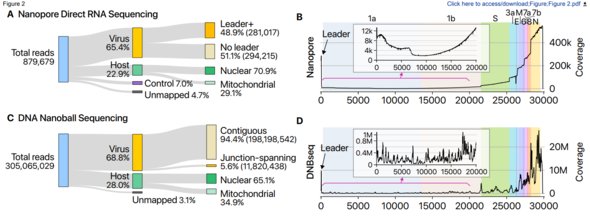
A COVID-19 infection begins when SARS-CoV-2, after infiltrating a host cell, copies its genomic RNA and produces a range of subgenomic RNA. Previous studies had only predicted gene location based on information about genomic RNA.
This study confirmed that only nine of the previously predicted ten subgenomes actually exist and identified a variety of chemical modifications that could present new characteristics. This helps researchers understand the coronavirus’s life cycle and pathogenicity, which could be used in developing a new therapeutic strategy.
“The newly discovered RNA and RNA modification sites represent potential targets in the development of viral medications,” V. Narry Kim said.
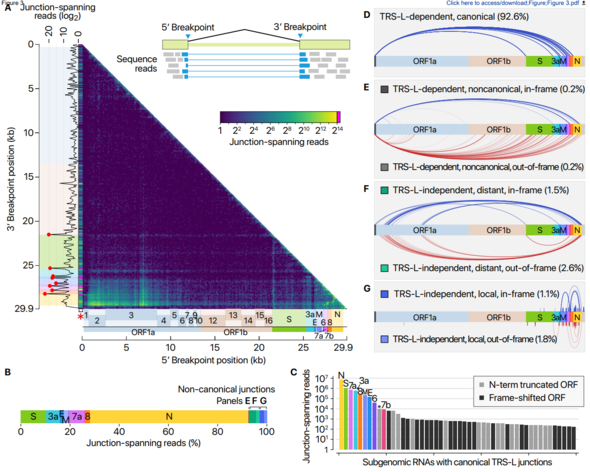
“Now that we’ve accurately determined the quantity of each transcriptome in SARS-CoV-2, that could serve as the basis for improving the polymerase chain reaction technique used to diagnose the disease,” added Chang Hye-shik.
This study applied the nanopore-based direct RNA sequencing approach (a next-generation technique for analyzing base sequences that was used for the first time in South Korea) to inactivated viruses provided by the KCDC. This new approach makes it possible to analyze SARS-CoV-2’s lengthy RNA sequences directly, without chopping them up. Under the typical approach, RNA is converted to DNA for analysis.
This paper was published online in Cell, an authoritative journal in the field of biology, on Apr. 9.
By Kim Jeong-su, senior staff writer
Please direct comments or questions to [english@hani.co.kr]

Editorial・opinion
![[Editorial] Perilous stakes of Trump’s rhetoric around US troop pullout from Korea [Editorial] Perilous stakes of Trump’s rhetoric around US troop pullout from Korea](https://flexible.img.hani.co.kr/flexible/normal/500/300/imgdb/original/2024/0509/221715238827911.jpg) [Editorial] Perilous stakes of Trump’s rhetoric around US troop pullout from Korea
[Editorial] Perilous stakes of Trump’s rhetoric around US troop pullout from Korea![[Guest essay] Preventing Korean Peninsula from becoming front line of new cold war [Guest essay] Preventing Korean Peninsula from becoming front line of new cold war](https://flexible.img.hani.co.kr/flexible/normal/500/300/imgdb/original/2024/0507/7217150679227807.jpg) [Guest essay] Preventing Korean Peninsula from becoming front line of new cold war
[Guest essay] Preventing Korean Peninsula from becoming front line of new cold war- [Column] The state is back — but is it in business?
- [Column] Life on our Trisolaris
- [Editorial] Penalties for airing allegations against Korea’s first lady endanger free press
- [Editorial] Yoon must halt procurement of SM-3 interceptor missiles
- [Guest essay] Maybe Korea’s rapid population decline is an opportunity, not a crisis
- [Column] Can Yoon steer diplomacy with Russia, China back on track?
- [Column] Season 2 of special prosecutor probe may be coming to Korea soon
- [Column] Park Geun-hye déjà vu in Yoon Suk-yeol
Most viewed articles
- 1Nuclear South Korea? The hidden implication of hints at US troop withdrawal
- 2‘Free Palestine!’: Anti-war protest wave comes to Korean campuses
- 3In Yoon’s Korea, a government ‘of, by and for prosecutors,’ says civic group
- 4[Photo] ‘End the genocide in Gaza’: Students in Korea join global anti-war protest wave
- 5[Editorial] Perilous stakes of Trump’s rhetoric around US troop pullout from Korea
- 6Seoul getting its first-ever vertical farm
- 7Gangnam murderer says he killed “because women have always ignored me”
- 8Real-life heroes of “A Taxi Driver” pass away without having reunited
- 9[Guest essay] Preventing Korean Peninsula from becoming front line of new cold war
- 10S. Korean first lady likely to face questioning by prosecutors over Dior handbag scandal